The
Hierarchical Data Format (HDF5) is a open-source, cross-platform,
data storage solution originally developed at the National Center for Supercomputing Applications (NCSA) for storage of scientific data
and descriptive meta-data in an organised heirarchy. This project extends the power of HDF5 to LabVIEW,
which is potentially very useful in many applications.
This project provides
HDF5 v1.10.4
functionality to
LabVIEW 2010 or newer, and is available
as a self-installing package (*.vip file), installable through the
VI Package Manager,
or the latest version can be installed
directly from the repository.
The h5labview library has been stable for many years, providing direct access to HDF data I/O functionality. However,
development of new functionality has ceased and the newer features of the HDF5 format may not be supported.
Users are welcome to implement any missing functionality.
News alert: Support for direct read and write of analog waveforms available as of v2.13.0
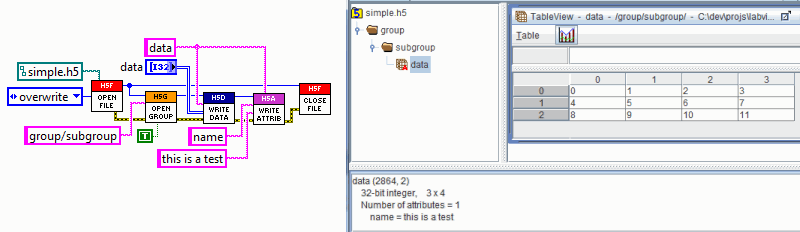
To demonstrate how easy it is to use
h5labview, here's an example showing creation of a new HDF5 file and writing
a dataset with an attribute inside a nested group. A screenshot from HDFView showing the hierarchy of the generated file
and the dataset is shown to the right.
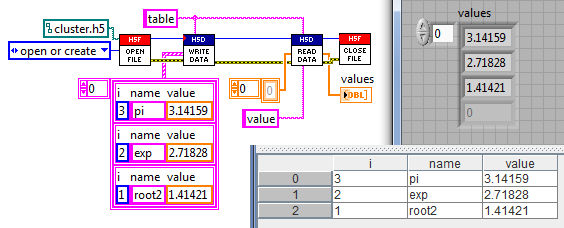
As of version 2,
h5labview supports compound datatypes (clusters) with "table" access, as demonstrated below.
The table below lists the datatypes supported by
h5labview in version 2, and their state of implementation.
See
this FAQ entry on why not all types are planned to be supported.
| LabVIEW type | HDF type | Notes |
| I8..I64 | H5T_NATIVE_INT8 .. H5T_NATIVE_INT64 | Native-endian numerical formats |
| U8..U64 | H5T_NATIVE_UINT8 .. H5T_NATIVE_UINT64 |
| SGL, DBL | H5T_NATIVE_FLOAT, H5T_NATIVE_DOUBLE |
| CSG, CDB | H5T_COMPOUND | Compound with "real" and "imag" components |
| EXT, CEX | Custom H5T_FLOAT | Host-compatible extended precision type1 |
| Physical quantities | Committed H5T_FLOAT | Written in SI units as named compounds |
| U8..U64 | H5T_BITFIELD | Read-only support for BITFIELD datatypes as unsigned integers |
| String | H5T_STRING | Both variable and fixed strings2 |
| Boolean | H5T_ENUM | Enumeration of "false"=0 and "true"=1 |
| Clusters | H5T_COMPOUND | Valid clusters may contain any of the above types |
| Arrays | Implemented as dataspaces3 | Array can be of any dimension of above type, including clusters |
| Enums | H5T_ENUM | LabVIEW enums supported4 |
| Images | As per specification | Writing of Grayscale, Truecolor and Palette images supported |
| Timestamps | H5T_COMPOUND | Direct translation of LabVIEW's internal timestamp format |
| Waveforms | Dataset with specific attributes | Currently only analog waveforms supported |
Notes:
- LabVIEW's memory layout for EXT varies across platforms, as does HDF's extended H5T_NATIVE_LDOUBLE.
A custom HDF type is created corresponding to the EXT layout on the host machine and HDF performs conversions automatically.
This should lead to no precision loss when writing, and enables reading at the greatest precision of the host.
The procedure is detailed in this FAQ entry.
- HDF supports two types of strings: fixed and variable length. Variable-length strings are implemented and used by default, but LabVIEW
strings that contain the null character (\0) must be written as fixed strings. Whether a string is read/written as fixed or variable
is configured in LabVIEW, as described in this FAQ entry.
- Arrays are implemented in HDF5 with a separate "dataspace" interface. Although an array datatype does exists, the H5T_ARRAY is inefficient
and serves limited purpose. The array datatype therefore will not be implemented, neither will arrays of arrays nor clusters including arrays.
- LabVIEW defines enums as a set of contiguous unsigned integers. HDF5 sets no such restriction, so as such any LabVIEW enum can be written
to an HDF file but HDF5 enums must be read as (signed) integers.


